The fate of a polygenic phenotype within the genomic landscapes of introgression in the European seabass hybrid zone
Unraveling the evolutionary mechanisms and consequences of hybridization is a major concern in biology. Many studies have documented the interplay between recombination and selection in modulating the genomic landscape of introgression, but few have considered how associations with phenotype may affect this landscape. Here, we use the European seabass (Dicentrarchus labrax), a key species in marine aquaculture that undergoes natural hybridization, to determine how selection on phenotype modulates the introgression landscape between Atlantic and Mediterranean lineages. We use a high-density SNP array to assess individual local ancestry along the genome and improve the mapping of muscle fat content, a polygenic trait that is divergent between lineages. Taking into account variation in recombination rates, we reveal a purging of Atlantic ancestry in the admixed Mediterranean populations. While Atlantic individuals had higher muscle fat content, we observed that genomic regions associated with this trait also displayed reduced introgression of Atlantic ancestry. These results emphasize how selection against maladapted alleles shape the genomic landscape of introgression.
Simple
- Date (Publication)
- 2020
- Date (Revision)
- 2024-10-03
- Other citation details
- Leitwein Maeva, Durif Ghislain, Delpuech Emilie, Gagnaire Pierre-Alexandre, Ernande Bruno, Vandeputte Marc, Vergnet Alain, Duranton Maud, Clota Frederic, Allal Francois (2020). The fate of a polygenic phenotype within the genomic landscapes of introgression in the European seabass hybrid zone. SEANOE. https://doi.org/10.17882/98771
- Credit
- L.M. was supported by a grant from Ifremer’s Scientific Board. This project was carried out with the support of the Ifremer MARBEC Experimental Aquaculture Research Station staff and facilities. This study received funding from European Union’s Horizon 2020 work program under the grant No. 652831 agreement (AQUAEXCEL2020), and from the French Ministry of the Sea (Direction générale des affaires maritimes, de la pêche et de l'aquaculture) under grant CRECHE2020.
Author
UMR Marbec, Université Montpellier, CNRS, Ifremer, IRD, INRAE, Montpellier, France
-
LEITWEIN Maeva
Author
UMR Marbec, Université Montpellier, CNRS, Ifremer, IRD, INRAE, Montpellier, France
-
DELPUECH Emilie
Author
UMR Marbec, Université Montpellier, CNRS, Ifremer, IRD, INRAE, Montpellier, France
-
ERNANDE Bruno
Author
UMR Marbec, Université Montpellier, CNRS, Ifremer, IRD, INRAE, Montpellier, France
-
VANDEPUTTE Marc
Author
UMR Marbec, Université Montpellier, CNRS, Ifremer, IRD, INRAE, Montpellier, France
-
VERGNET Alain
Author
UMR Marbec, Université Montpellier, CNRS, Ifremer, IRD, INRAE, Montpellier, France
-
DURANTON Maud
Author
UMR Marbec, Université Montpellier, CNRS, Ifremer, IRD, INRAE, Montpellier, France
-
CLOTA Frederic
Author
UMR Marbec, Université Montpellier, CNRS, Ifremer, IRD, INRAE, Montpellier, France
-
ALLAL Francois
- Theme
-
- Hybridization
- Introgression
- phenotype
- European seabass
- selection
- recombination
- Environment
- project
-
- FP7 AQUA EXCEL (Agreement: 262336)
- Use constraints
- Other restrictions
- Date (Publication)
- 2024
- Unique resource identifier
- 10.1093/molbev/msae194
- Association Type
- Cross reference
- Initiative Type
- Study
- Metadata language
- English
- Topic category
-
- Oceans
N
S
E
W
))
- Distribution format
-
-
VCF
(
)
-
VCF
(
)
- OnLine resource
-
Raw data
(
WWW:DOWNLOAD-1.0-link--download
)
Genotyping data in VCF format for Offspring and parents - 353 MB
- OnLine resource
- DOI of the product ( WWW:LINK-1.0-http--metadata-URL )
- OnLine resource
- Seanoe ( rel-canonical )
- Hierarchy level
- Dataset
- Statement
- Genotyping data obtained by DLabCHIP Axiom SNP array (Griot et al, 2020, https://doi.org/10.1016/j.aquaculture.2020.735930)
- File identifier
- seanoe:98771 XML
- Metadata language
- English
- Character set
- UTF8
- Hierarchy level
- Dataset
- Date stamp
- 2024-10-03
- Metadata standard name
- ISO 19115:2003/19139
- Metadata standard version
- 1.0
Overviews
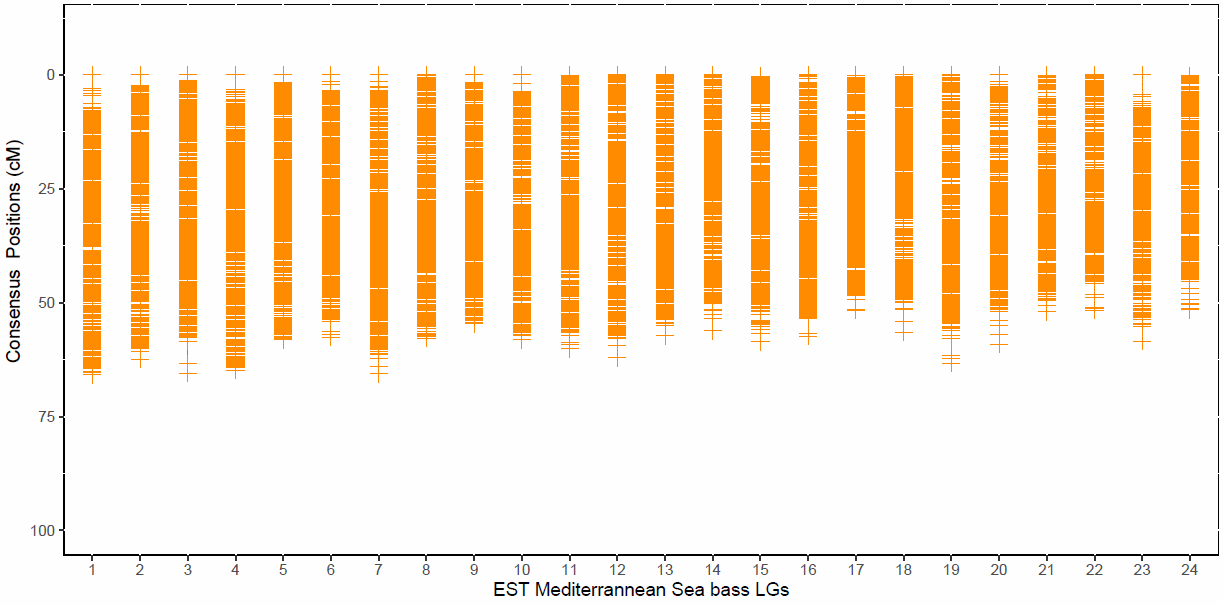
Genetic card obtained for the offspring of the study from SNP genetyped with the DLabCHIP Axiom array
Spatial extent
N
S
E
W
))
Provided by

Associated resources
Not available
 Catalogue PIGMA
Catalogue PIGMA