DOC, DOCX
Type of resources
Available actions
Topics
Keywords
Contact for the resource
Provided by
Years
Formats
-
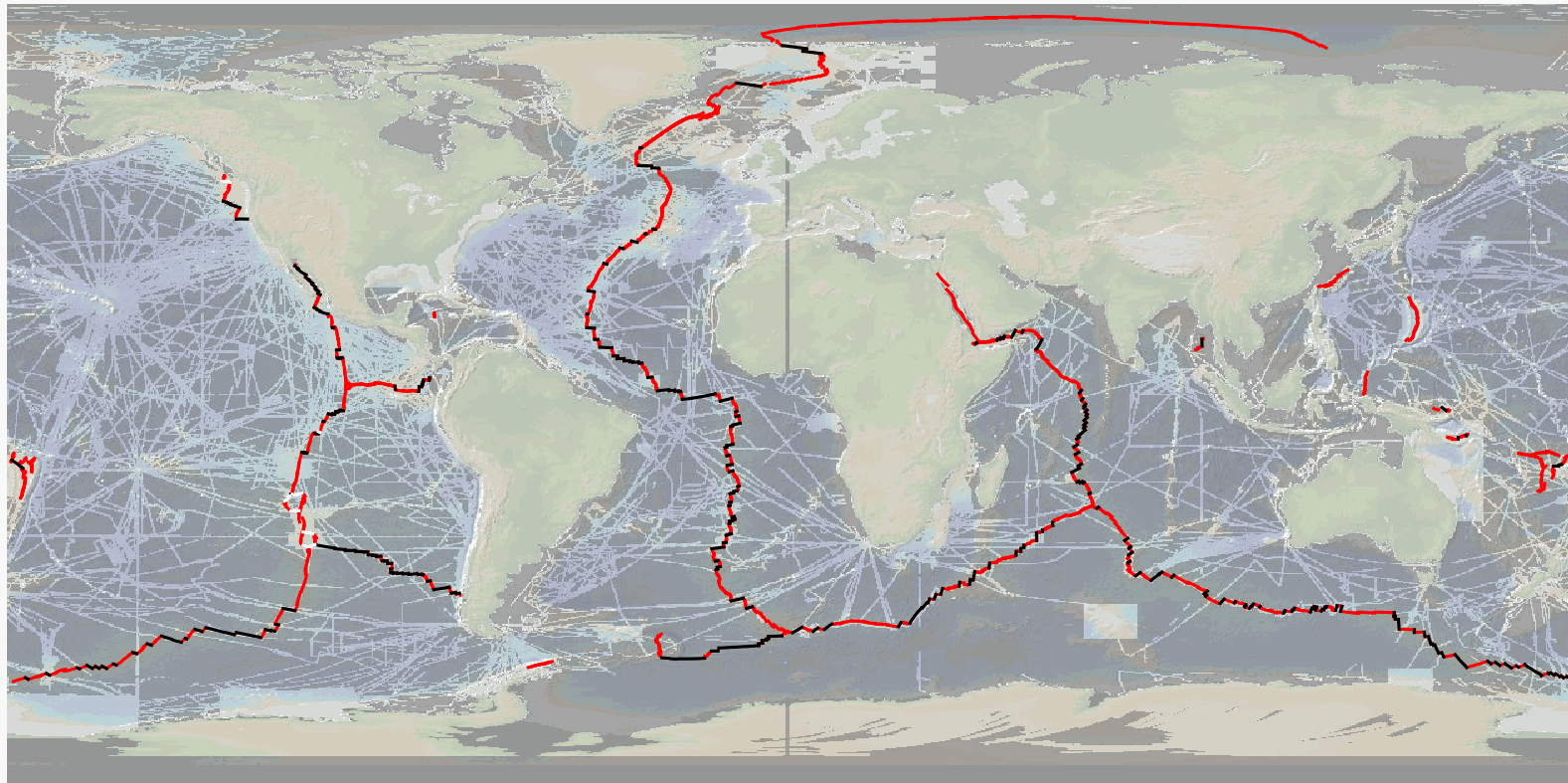
The precise location and geometry of oceanic spreading centers and associated transform faults or discontinuities' boundary has fundamental implications in our understanding of oceanic accretion, the accommodation of deformation around rigid lithospheric blocks, and the distribution of magmatic and volcanic processes. The now widely used location of mid oceanic ridges worldwide, published by P. Bird in 2003, can be updated based on recent publicly available and published ship-based multibeam swath bathymetry data (100-m resolution or better), now available to ~25% of the ocean seafloor, but covering a significant proportion of the mid-ocean ridge system (>70%). Here we publish the MAPRIDGES database built under the coordination of CGMW (Commission for the Geological Map of the World), with a first version V1.0 (06/2024) that provides high resolution and up-to-date datasets of mid-ocean ridge segments and associated transform faults, and follow-up updates that will also include non-transform offsets. The detailed mapping of individual mid oceanic ridge segments was conducted using GMRT (Ryan et al., 2009) (version 4.2 for MAPRIDGES V1.0), other publicly available datasets (e.g., NCEI, Pangaea, AWI), and existing literature. MAPRIDGES will be revised with the acquisition of additional datasets, new publications, and correction of any errors in the database. The MAPRIDGE database was built in a GIS environment, where each feature holds several attributes specific to the dataset. We include three different georeferenced shapefile layers: 1) Ridge Segments, 2) Transform Faults, and 3) Transform Zones. The latest corresponds to zones of distributed strike-slip deformation that lack a well-defined fault localizing strain, but that are often treated as transform faults. 1) The Ridge Segments Layer contains 1461 segments with 9 attributes: - AREA_LOCA: The Name of the Ridge System - LOC_SHORT: The short form of the Ridge System using 3 characters - LAT: The maximum latitude of the ridge segment - LONG: the maximum longitude of the ridge segment - LENGTH: the length of the ridge segment in meters - CONFIDENCE: the degree of confidence on digitization based on the availability of high-resolution bathymetry data: 1 = low to medium confidence, 2 = high confidence - REFERENCES: supporting references used for the digitization - NAME_CODE: unique segment code constructed from the LOC_SHORT and LAT attributes in degree, minute, second coordinate format - NAME_LIT: name of the segment from the literature if it exists 2) The Transform Fault Layer contains 260 segments with 4 attributes: - NAME_TF: Name of the transform fault according to the literature - LENGTH: length of the transform fault in meters - LAT: The maximum latitude of the fault segment - LONG: the maximum longitude of the fault segment 3) The Transform Zone layer contains 10 segments with 4 attributes: - NAME_TF: Name of the transform zone according to the literature - LENGTH: length of the transform fault in meters - LAT: The maximum latitude of the fault segment - LONG: the maximum longitude of the fault segment To facilitate revisions and updates of the database, relevant information, corrections, or data could be sent to B. Sautter (benjamin.sautter@univ-ubs.fr) and J. Escartín (escartin@geologie.ens.fr).
-

The present repository makes available the model, material and outputs of the ISIS-Fish modeling work showcased in the peer-reviewed scientific article by Bastardie et al. 2025. As part of the SEAwise research project (seawiseproject.org), we used an ISIS-Fish database (Mahevas et al 2003, Pelletier et al. 2009, isis-fish.org) previously developed within the MACCO project which describes the mixed demersal fishery in the Bay of Biscay. For this application, the spatial extent of the fishery is the Bay of Biscay, defined here by ICES divisions 8a, 8b and 8d and the resolution chosen is 1/16 ICES statistical rectangle. The biological module (Vajas et al. 2024) includes 7 species of economic interest in the mixed demersal fishery: European hake (Merluccius merluccius), common sole (Solea solea), Norway lobster (Nephrops norvegicus), megrim (Lepidorhombus whiffiagonis), anglerfish (Lophius piscatorius) and two ray species (Raja clavata, Leucoraja naevus). The fishing activities module (Mahevas et al. 2024) is made up of 41 demersal fleets (including all French vessels < 12 meters and > 12 meters fishing in this area, Spanich, UK and Belgium fleets) and 431 métiers (combination of a gear, location and mix of target species) catching these 7 species, as target or bycatch. Monthly effort of a fleet distributes among the possible métiers (those historically practiced). The biological and fishing activity modules are identical to the published version. The original model used here has been calibrated on historical catch data 2015-2018 by tuning accessibility and catchability parameters. In the present application the Bay of Biscay model is used to investigate the spatial- and effort- based fisheries management strategies. Consistently with for a task of the SEAwise project (Bastardie et al. 2024) simulations were conducted from 2021 onwards, projecting the effect of an implementation of 3 different closures from 2022 to 2050, under current fishing effort conditions or in a context of fishing effort reduction. Outcomes of these simulations are averaged over short/medium (10 year horizon) and long-term period (20 year horizon). The data project includes: 1) the database including the biological module and fishing activity module; 2) 8 .properties files, each corresponding to one combination of management measure and closure, to restore the simulations parameters in the ISIS-Fish interface and reproduce the simulation runs; 3) the .java scripts to force effort dynamics and simulate spatio-temporal closures, as well as generate the main output files - they will be called by the ISIS-Fish software once the simulations restored 4) the .rds containing the main outputs of the simulations and the associated .html document displaying the R code to compute the indices of interest at different levels of aggregation and reproduce the figures in Bastardie et al. 2025. All files are provided in the Zip. Associated with this material, a study summary and a readme .docx are provided. The first one provides context on the present work and describes the model and simulations' design. The second provides guidelines to reproduce the simulations and their derived outcomes from the data project material made available in this repository. They are both directly downloadable from this repository and are also copied to the zipped folder containing the data project. All the data are reproducible using isis-fish-4.4.8.1 (isis-fish.org; available at forge.codelutin.com) and R 4.2.0.
 Catalogue PIGMA
Catalogue PIGMA