2023
Type of resources
Available actions
Topics
Keywords
Contact for the resource
Provided by
Years
Formats
Representation types
Update frequencies
status
Scale
Resolution
-
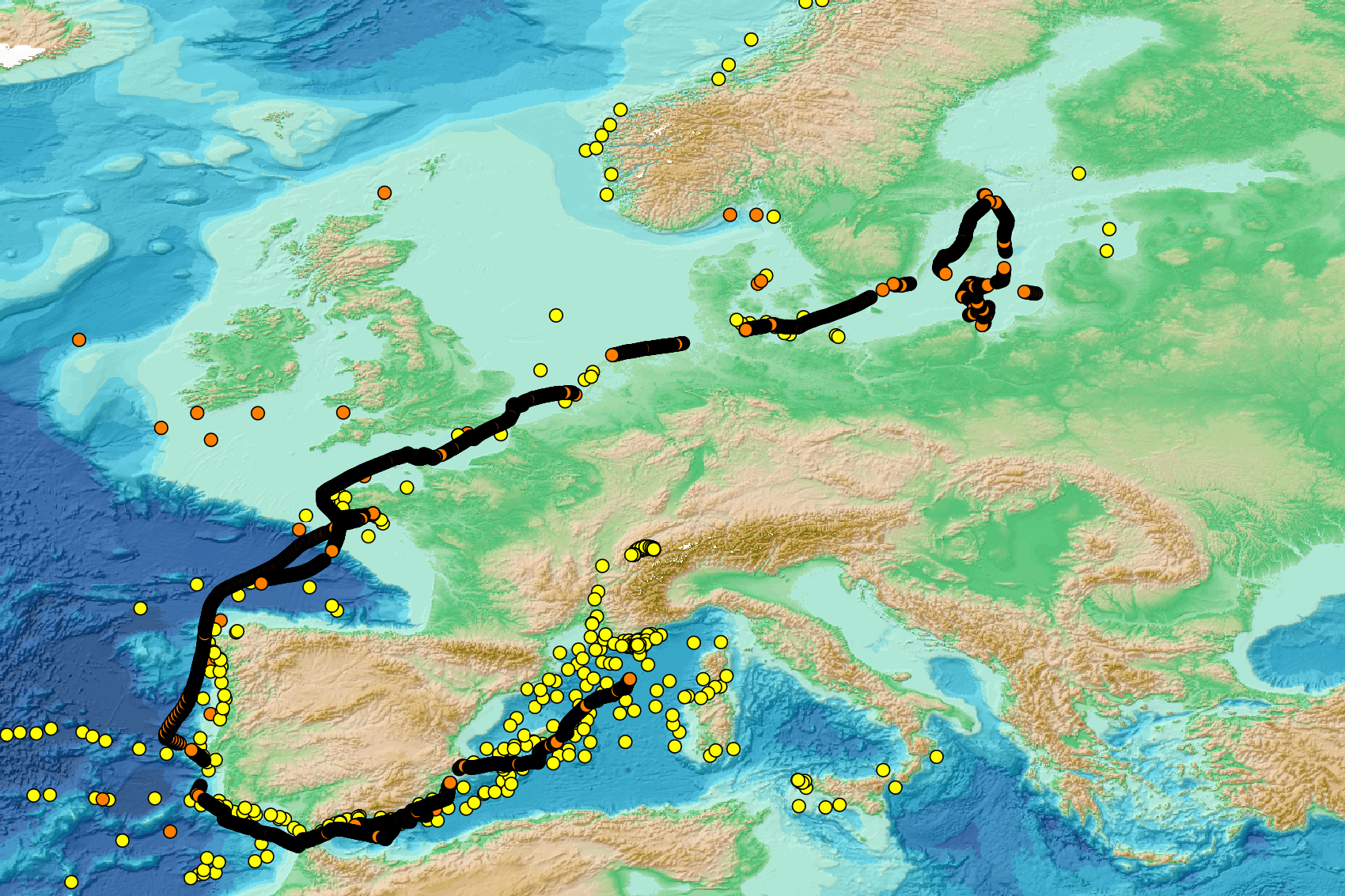
This visualization product displays the location of all the surveys present in the EMODnet marine litter database (MLDB). The different fishing gears used are represented by different colors. EMODnet Chemistry included the collection of marine litter in its 3rd phase. Since the beginning of 2018, data of seafloor litter collected by international fish-trawl surveys have been gathered and processed in the EMODnet Chemistry Marine Litter Database (MLDB). The harmonization of all the data has been the most challenging task considering the heterogeneity of the data sources, sampling protocols (OSPAR and MEDITS protocols) and reference lists used on a European scale. Moreover, within the same protocol, different gear types are deployed during fishing bottom trawl surveys. Unlike other EMODnet seafloor litter products, all trawls surveyed since 2006 are included in this map even if the wingspread and/or the distance are unknown. Only surveys with an unknown number of items were excluded from this product. More information on data processing and calculation are detailed in the document attached. Warning: the absence of data on the map doesn't necessarily mean that they don't exist, but that no information has been entered in the Marine Litter Database for this area.
-

Species distribution models (GAM, Maxent and Random Forest ensemble) predicting the distribution of Solitary Scleractinian fields assemblage in the Celtic Sea. This community is considered ecologically coherent according to the cluster analysis conducted by Parry et al. (2015) on image sample. Modelling its distribution complements existing work on their definition and offers a representation of the extent of the areas of the north-east Atlantic where they can occur based on the best available knowledge. This work was performed at the University of Plymouth in 2021.
-
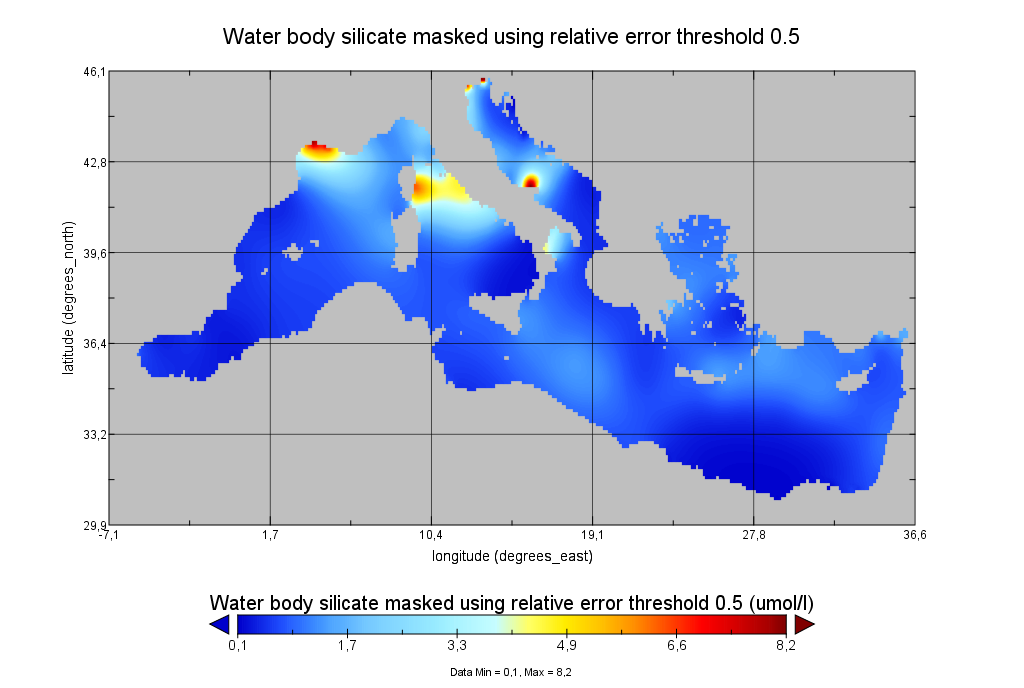
Moving 6-year analysis of Water body silicate in the Mediterranean Sea for each season: - winter: January-March, - spring: April-June, - summer: July-September, - autumn: October-December. Every year of the time dimension corresponds to the 6-year centered average of the season. 6-years periods span from 1970-1975 until 2017-2022. Data Sources: observational data from SeaDataNet/EMODNet Chemistry Data Network. Units: umol/l. Description of DIVA analysis: The computation was done with the DIVAnd (Data-Interpolating Variational Analysis in n dimensions), version 2.7.9, using GEBCO 30sec topography for the spatial connectivity of water masses. The horizontal resolution of the produced DIVAnd maps grids is dx=dy=0.125 degrees (around 13.5km and 10.9km accordingly). The vertical resolution is 25 depth levels: [0.,5.,10.,20.,30.,50.,75.,100.,125.,150.,200.,250.,300.,400.,500.,600.,700.,800.,900.,1000.,1100.,1200.,1300.,1400.,1500.]. The horizontal correlation length is 200km. The vertical correlation length (in meters) was set twices the vertical resolution: [10.,10.,20.,20.,40.,50.,50.,50.,50.,100.,100.,100.,200.,200.,200.,200.,200.,200.,200.,200.,200.,200.,200.,200.,200.]. Duplicates check was performed using the following criteria for space and time: dlon=0.001deg., dlat=0.001deg., ddepth=1m, dtime=1hour, dvalue=0.1. The error variance (epsilon2) was set equal to 1 for profiles and 10 for time series to reduce the influence of close data near the coasts. An anamorphosis transformation was applied to the data (function DIVAnd.Anam.loglin) to avoid unrealistic negative values: threshold value=200. A background analysis field was used for all years (1970-2022) with correlation length equal to 600km and error variance (epsilon2) equal to 20. Quality control of the observations was applied using the interpolated field (QCMETHOD=3). Residuals (differences between the observations and the analysis (interpolated linearly to the location of the observations) were calculated. Observations with residuals outside the minimum and maximum values of the 99% quantile were discarded from the analysis. Originators of Italian data sets-List of contributors: - Brunetti Fabio (OGS) - Cardin Vanessa, Bensi Manuel doi:10.6092/36728450-4296-4e6a-967d-d5b6da55f306 - Cardin Vanessa, Bensi Manuel, Ursella Laura, Siena Giuseppe doi:10.6092/f8e6d18e-f877-4aa5-a983-a03b06ccb987 - Cataletto Bruno (OGS) - Cinzia Comici Cinzia (OGS) - Civitarese Giuseppe (OGS) - DeVittor Cinzia (OGS) - Giani Michele (OGS) - Kovacevic Vedrana (OGS) - Mosetti Renzo (OGS) - Solidoro C.,Beran A.,Cataletto B.,Celussi M.,Cibic T.,Comici C.,Del Negro P.,De Vittor C.,Minocci M.,Monti M.,Fabbro C.,Falconi C.,Franzo A.,Libralato S.,Lipizer M.,Negussanti J.S.,Russel H.,Valli G., doi:10.6092/e5518899-b914-43b0-8139-023718aa63f5 - Celio Massimo (ARPA FVG) - Malaguti Antonella (ENEA) - Fonda Umani Serena (UNITS) - Bignami Francesco (ISAC/CNR) - Boldrini Alfredo (ISMAR/CNR) - Marini Mauro (ISMAR/CNR) - Miserocchi Stefano (ISMAR/CNR) - Zaccone Renata (IAMC/CNR) - Lavezza, R., Dubroca, L. F. C., Ludicone, D., Kress, N., Herut, B., Civitarese, G., Cruzado, A., Lefèvre, D.,Souvermezoglou, E., Yilmaz, A., Tugrul, S., and Ribera d'Alcala, M.: Compilation of quality controlled nutrient profiles from the Mediterranean Sea, doi:10.1594/PANGAEA.771907, 2011.
-
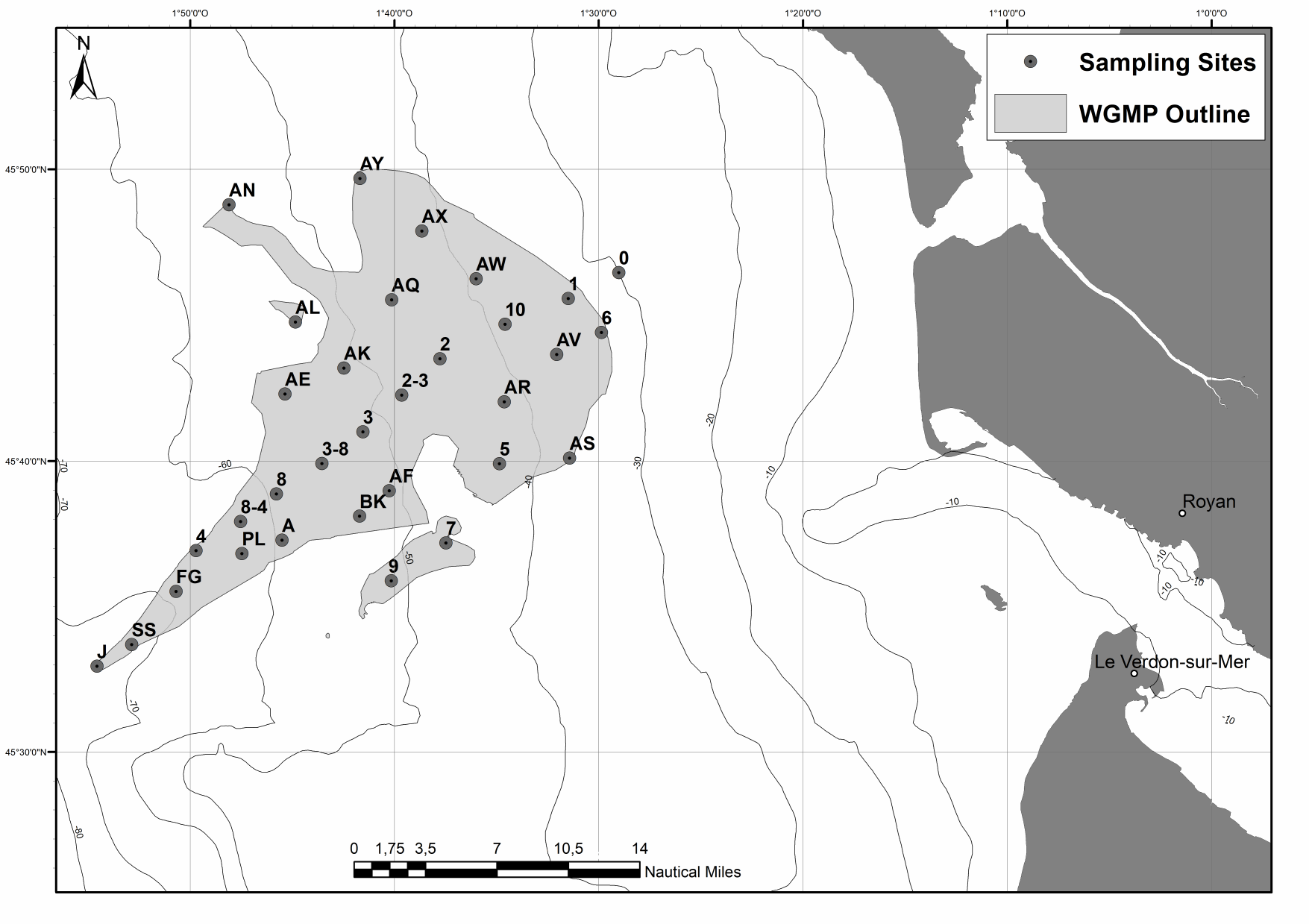
The spatial distributions of (1) surface sediment characteristics (D0.5, Sediment Surface Area (SSA), Particulate Organic Carbon (POC), Chlorophyll-a (Chl-a), Phaeophytin-a (Phaeo-a), Total and Enzymatically Hydrolyzable Amino Acids (THAA, EHAA), δ13C) and (2) sediment profile image (apparent Redox Potential Discontinuity (aRPD), numbers and depths of biological traces) characteristics were quantified based on the sampling of 32 stations located within the West Gironde Mud Patch (Bay of Biscay, NE Atlantic) in view of (1) assessing the spatial structuration of a temperate river-dominated ocean margin located in a high-energy area, (2) disentangling the impacts of hydrodynamics and bottom trawling on this structuration, and (3) comparing the West Gironde Mud Patch with the Rhône River Prodelta (located in a low-energy area). Results support the subdivision of the West Gironde Mud Patch in a proximal and a distal part and show (1) the existence of depth gradients in surface sedimentary organics characteristics and bioturbation within the distal part; (2) no evidence for a significant effect of bottom trawling, as opposed to Bottom Shear Stress, on the West Gironde Mud Patch spatial structuration; and (3) major discrepancies between spatial structuration in the West Gironde Mud Patch and the Rhône River Prodelta, which were attributed to differences in tidal regimes, sedimentation processes, and local hydrodynamics, which is in agreement with current river-dominated ocean margin typologies.
-
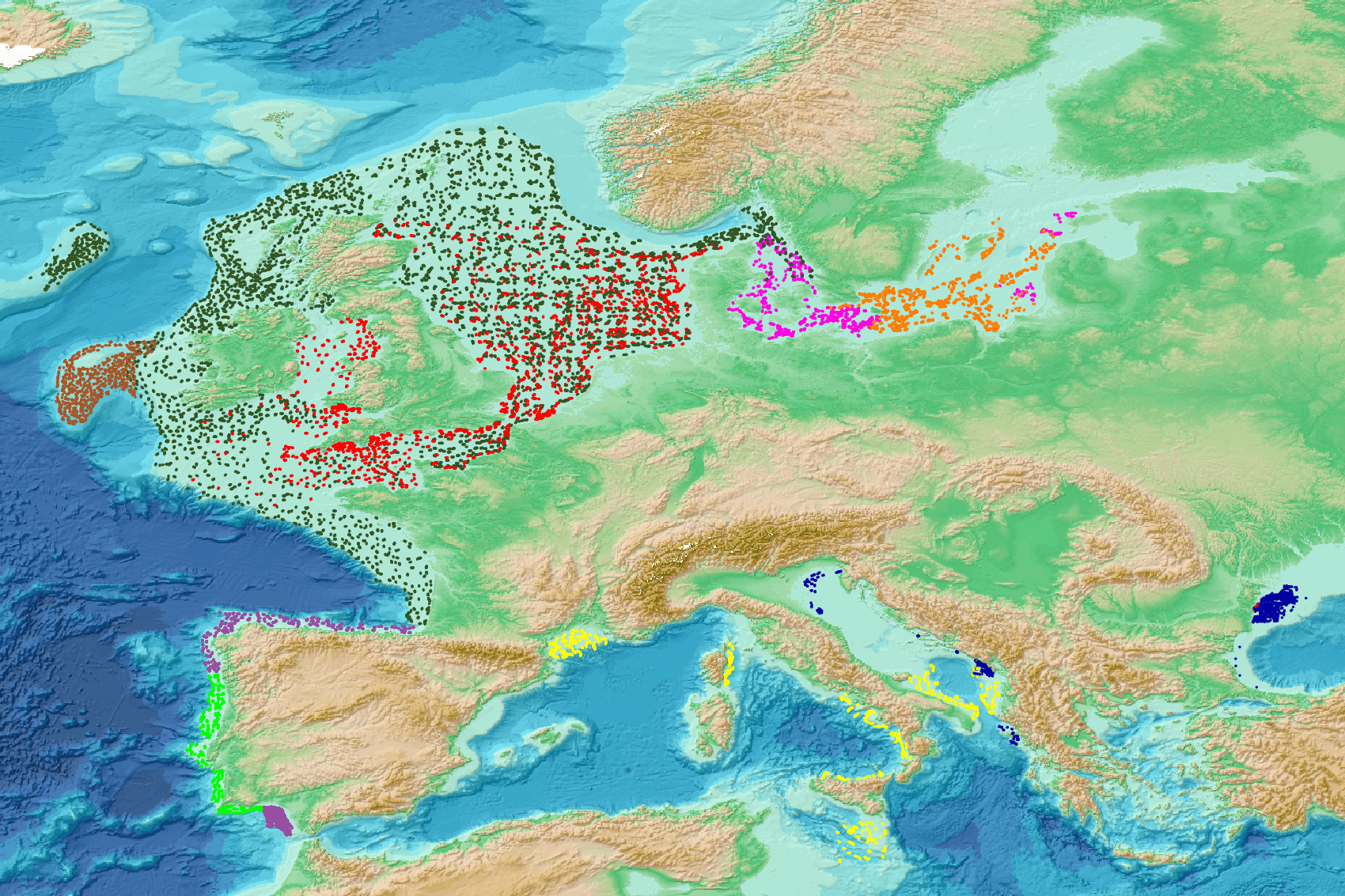
This visualization product displays nets locations where specific protocols have been applied to collate data on microlitter. Mesh size used with these protocols have been indicated with different colors in the map. EMODnet Chemistry included the collection of marine litter in its 3rd phase. Before 2021, there was no coordinated effort at the regional or European scale for micro-litter. Given this situation, EMODnet Chemistry proposed to adopt the data gathering and data management approach as generally applied for marine data, i.e., populating metadata and data in the CDI Data Discovery and Access service using dedicated SeaDataNet data transport formats. EMODnet Chemistry is currently the official EU collector of micro-litter data from Marine Strategy Framework Directive (MSFD) National Monitoring activities (descriptor 10). A series of specific standard vocabularies or standard terms related to micro-litter have been added to SeaDataNet NVS (NERC Vocabulary Server) Common Vocabularies to describe the micro-litter. European micro-litter data are collected by the National Oceanographic Data Centres (NODCs). Micro-litter map products are generated from NODCs data after a test of the aggregated collection including data and data format checks and data harmonization. A filter is applied to represent only micro-litter sampled according to a very specific protocol such as the Volvo Ocean Race (VOR) or Oceaneye. Warning: the absence of data on the map doesn't necessarily mean that they don't exist, but that no information has been entered in the National Oceanographic Data Centre (NODC) for this area.
-
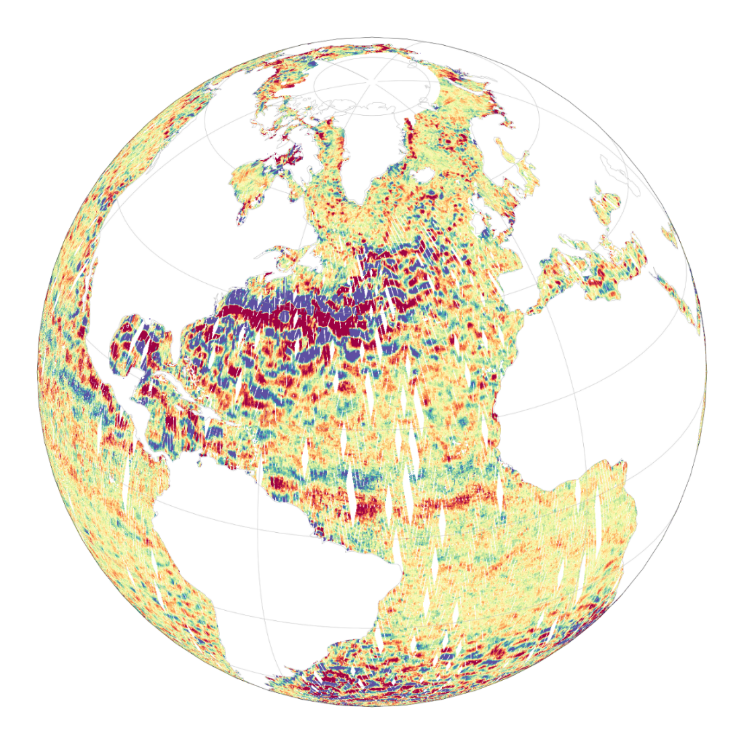
The Level-2 Ka-band Radar Interferometer (KaRIn) low rate (LR, ocean) sea surface height (SSH) data product from the Surface Water and Ocean Topography (SWOT) mission, also referenced by the short name L2_LR_SSH, provides ocean topography measurements from the low rate ocean data stream of the KaRIn instrument, spanning 60 km on either side of the nadir altimeter with a nadir gap. The L2_LR_SSH product is available continuously and globally, although different versions of the product may be produced at different latencies and/or through different reprocessing with refined input data. Note that L2_LR_SSH does not include SSH data from the SWOT nadir altimeter. The SWOT L2_LR_SSH product is organized as a collection of four subproducts, the L2_LR_SSH ['Basic'] is described in this metadata sheet. The 3 other file types (['WindWave'], ['Expert'], ['Unsmoothed']) are described by 3 different metadata sheets that can be accessed through the links below. The ['Basic'] file on this metadata sheet, is intended for users who are interested in SSH measurements and who will use the KaRIn measurements as provided. The ['Basic'] L2_LR_SSH includes corrected sea surface height (SSH), sea surface height anomaly (SSHA), data quality flags, geophysical reference fields, and height correction information on a 2-km geographically fixed grid. The ['WindWave'] L2_LR_SSH is intended for users interested in wind and wave information. The ['Expert'] L2_LR_SSH is intended for expert users who are interested in the details of how the KaRIn measurements were derived and who may use detailed information for their own custom processing. The ['Unsmoothed'] L2_LR_SSH, also intended for expert users, is provided on a finer 'native' grid of 250-m (with minimal smoothing applied), and has a significantly larger data volume than the other files. May 2025: v3.0 (version D) Production and distribution of the L2_LR_SSH version D products: - PID0 for forward-processed version D products: from May 6, 2025 onward, - PGD0 for reprocessed version D products: from March 30 to July 10, 2023 (phase CalVal) and from July 26, 2023 to May 19, 2025 (phase Science) is ongoing. August 2024: v2.0 (version D) L2_LR_SSH version C products declared as validated by the SWOT project. March 2024: v2.0 (version C) Production and distribution of the pre-validated L2_LR_SSH version C products: - PIC0 for forward-processed version C products: November 23, 2023 to present, - PGC0 for reprocessed version C products: from March 30 to July 10, 2023 (phase CalVal) and from July 26, 2023 to January 25, 2024 (phase Science) November 2023: v1.0 The beta pre-validated L2_LR_SSH version 1.0 product (summer 2023 reprocessing release) is available only for the 1-day CalVal orbit phase, from March 29 to July 10, 2023, and the 21-day Science orbit phase from September 7 to November 21, 2023.
-

This product displays for Cadmium, positions with percentages of all available data values per group of animals that are present in EMODnet regional contaminants aggregated datasets, v2022. The product displays positions for all available years.
-
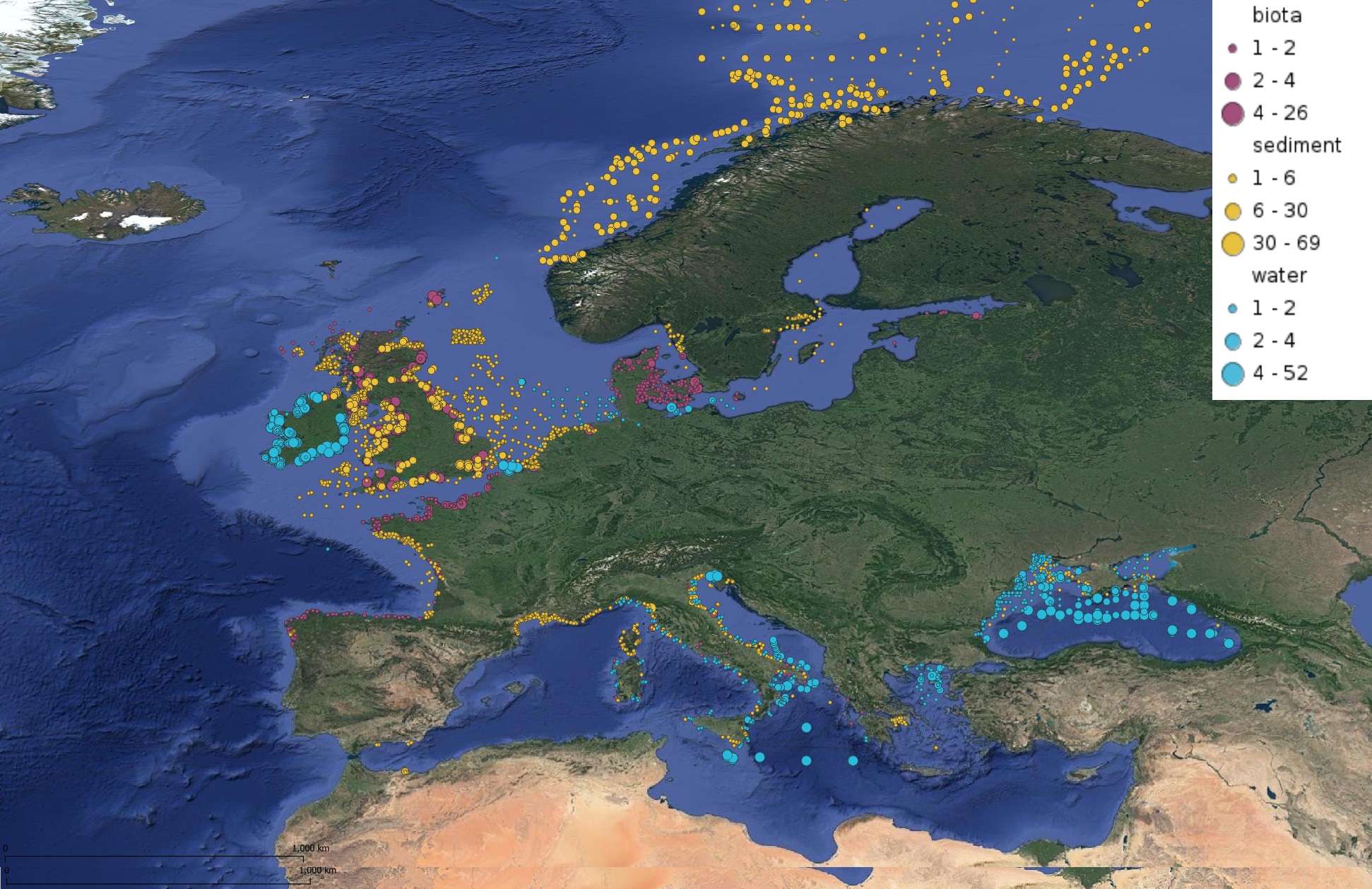
This product displays for Benzo(a)pyrene, positions with values counts that have been measured per matrix for each year and are present in EMODnet regional contaminants aggregated datasets, v2022. The product displays positions for every available year.
-
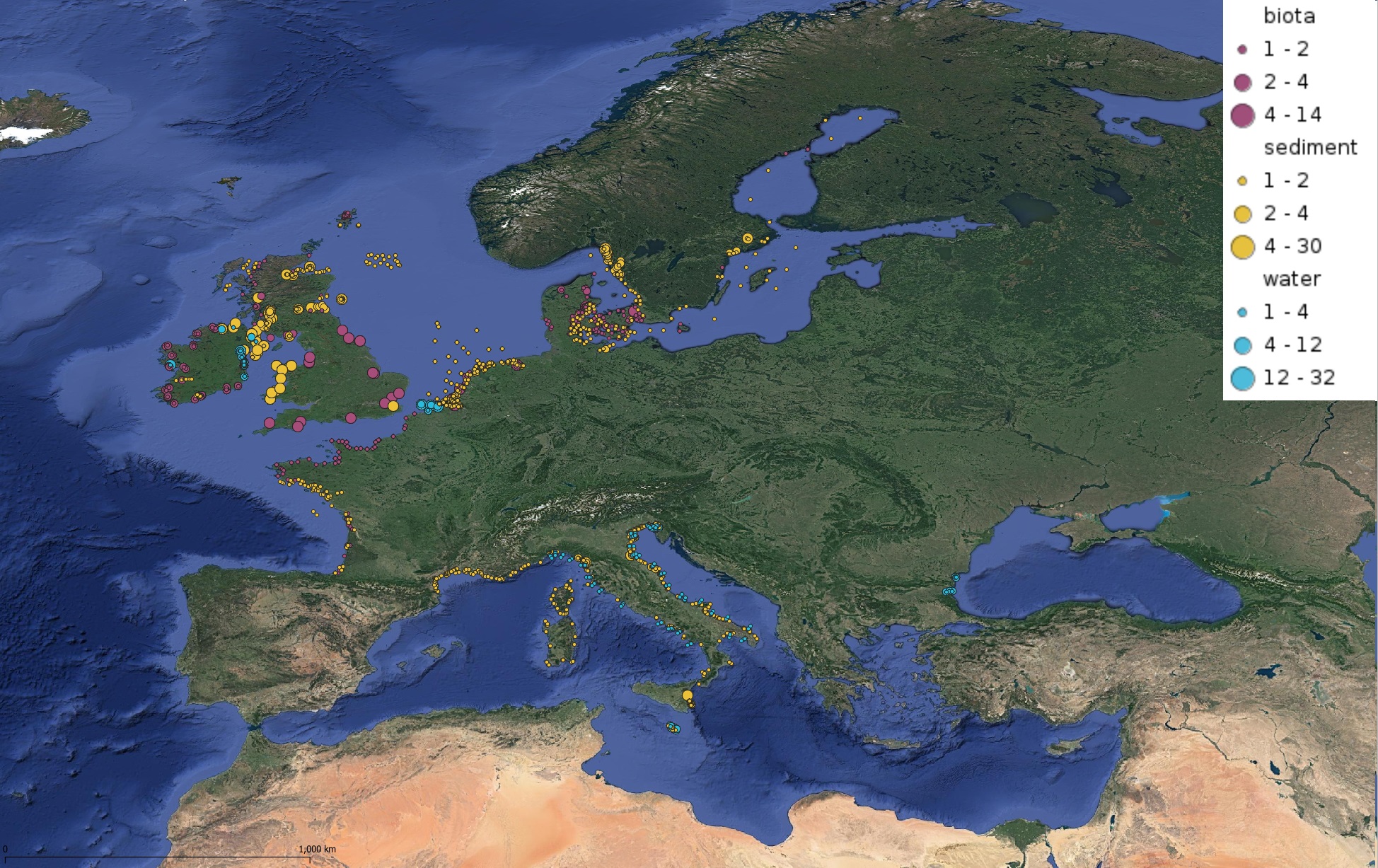
This product displays for Tributyltin, positions with values counts that have been measured per matrix for each year and are present in EMODnet regional contaminants aggregated datasets, v2022. The product displays positions for every available year.
-

In order to better characterize the fish biodiversity associated to the presence of cetaceans, especially the common Dolphin, from the Bay of Biscay, sequences from a 16S fragment were obtained from water samples acquired close to groups of dolphins.
 Catalogue PIGMA
Catalogue PIGMA