Phytoplankton
Type of resources
Available actions
Topics
Keywords
Contact for the resource
Provided by
Years
Formats
Representation types
Update frequencies
status
Scale
Resolution
-
The data-set is composed of three tables, Environmental variables, Phytoplankton ( in log+1 abundance) and the coordinates of the station used in the study. They are the processed data.
-

This dataset contains the biological outputs of a global ocean simulation coupling dynamics and biogeochemistry at ¼° over the year 2019. The simulation has been performed using the coupled circulation/ecosystem model NEMO/PISCES (https://www.nemo-ocean.eu/), which is here enhanced to perform an ensemble simulation with explicit simulation of modeling uncertainties in the physics and in the biogeochemistry. This dataset is one of the 40 members of the ensemble simulation. This study was part of the Horizon Europe project SEAMLESS (https://seamlessproject.org/Home.html), with the general objective of improving the analysis and forecast of ecosystem indicators. See Popov et al. (https://os.copernicus.org/articles/20/155/2024/) for more details on the study.
-
The dataset dcm_dtb.txt contains bio-optical measurements and environmental parameters associated with Deep Chlorophyll Maxima (DCM) acquired by BGC-Argo profiling floats. For each BGC-Argo profile the data files includes the World Meteorological Organization (WMO) and profile numbers, the Data Assembly Center (DAC), the geographical position (LON and LAT), the date of the profile in Julian Day (JULD) and in YYYY-MM-DD format; the region of the profile (REGION, acronyms detailed in the region.txt file), the DCM zonal attribution (ZONE, acronyms detailed in the zone.txt file), the vertical resolution of measurements of the concentration of the chlorophyll a [Chla] and of the backscattering coefficient (bbp) within the 250 first meters, the Mixed Layer Depth (MLD, m), the qualification of the vertical profile (DCM_TYPE) as Deep Biomass Maximum (3), Deep photoAcclimation Maximum (2), or presenting no DCM (1); the depth of the DCM (DCM_DEPTH); the chlorophyll a concentration (CHLA_DCM, mg chla m-3 ) the backscattering coefficient (BBP_DCM, m-1), and the Brunt-Vaisala frequency (N2_DCM) at the DCM depth; the nitracline depth (NCLINE_DEPTH, m) and steepness (NCLINE_STEEP, µmol NO3 m-3 m-1), the mean nitrate concentration within the Mixed Layer (NO3_MEAN_MLD, µmol NO3 m-3), the mean daily Photosynthetically Available Radiation in the Mixed Layer (MEAN_IPAR_MLD, E m -1 d -1), the daily Photosynthetically Available Radiation at the nitracline depth (IPAR_NCLINE, E m-2 d-1); and the [Chla] measured by satellite (CHLA_SAT, mg chla m-3). The dataset shape_NASTG_ASEW.txt contains the seasonal median, the first and third quartiles of the [Chla] and of the bbp profiles for the North Atlantic Subtropical Gyre and Atlantic SubEquatorial Waters regions. The dataset climato_NASTG_ASEW.txt contains the monthly mean and standard deviations of the DCM depth (DCM_depth), the isolume depth of daily Photosynthetically Available Radiation of 20 E m-2 d-1 (iPAR_20), the nitracline depth, and the Mixed Layer Depth (MLD) for the profiles within the North Atlantic Subtropical Gyre and Atlantic SubEquatorial Waters regions. The qualification and processing of the BGC-Argo profiles, as well as the DCM detection (DCM_TYPE) and the estimation of the environmental parameters, were applied as described from Cornec, M., Claustre, H., Mignot, A., Guidi, L., Lacour, L., Poteau, A., D’Ortenzio, F.,Gentili, B., Schmechtig, C., (to be updated.) Deep Chlorophyll Maxima in the global ocean: occurrences, drivers and characteristics. Global Biogeochemical Cycles, to be updated The [Chla] satellite variable was obtained by the match of each BGC-Argo profile with a L3S [Chla] product from the Ocean Colour-Climate Change Initiative v4.0 database merging observations from MERIS, MODIS, VIIRS and SeaWiFs, at a monthly and 4x4-km-pixel resolution, up to December 31, 2019 (ftp://oc-cci-data:ELaiWai8ae@oceancolour.org/occci-v4.2/).
-

'''Short description:''' Le modèle biogéochimique ECO-MARS3D sur la façade Manche Atlantique (PREVIMER_B1-ECOMARS3D-MANGA4000) est un modèle 3D de résolution spatiale 4km qui fournit les concentrations de nutriments et de plancton toutes les heures sur 30 niveaux (fenêtre de prévision à 4 jours). '''Paramètres calculés :''' Les paramètres calculés sont les suivants : * SAL : sea_water_salinity * TEMP : sea_water_temperature * suspended_inorganic_particulate_matter : mass_concentration_of_suspended_matter_in_sea_water * nanopicoplankton_nitrogen : mole_concentration_of_nanoplankton_expressed_as_nitrogen_in_sea_water * diatom_nitrogen : mole_concentration_of_diatoms_expressed_as_nitrogen_in_sea_water * dinoflagellate_nitrogen : mole_concentration_of_dinoflagellates_expressed_as_nitrogen_in_sea_water * microzooplankton_nitrogen : mole_concentration_of_microzooplankton_expressed_as_nitrogen_in_sea_water * mesozooplankton_nitrogen : mole_concentration_of_mesozooplankton_expressed_as_nitrogen_in_sea_water * colonial_phaeocystis_nitrogen : mole_concentration_of_colonial_phaeocystis_expressed_as_nitrogen_in_sea_water * phaeocystis_mucus : concentration_of_phaeocystis_mucus_expressed_as_mass_in_sea_water * ammonium : mole_concentration_of_ammonium_in_sea_water * nitrate : mole_concentration_of_nitrate_in_sea_water * dissolved_silicate : mole_concentration_of_silicate_in_sea_water * dissolved_phosphate : mole_concentration_of_phosphate_in_sea_water * dissolved_oxygen : dissolved_oxygen_in_water_column * cumulative_nanoflagellate_carbon_production : cumulative_nanoflagellate_production_expressed_as_carbon_in_sea_water * cumulative_diatom_carbon_production : cumulative_diatom_production_expressed_as_carbon_in_sea_water * cumulative_dinoflagellate_carbon_production : cumulative_dinoflagellate_production_expressed_as_carbon_in_sea_water * cumulative_phaeocystis_carbon_production : cumulative_phaeocystis_production_expressed_as_carbon_in_sea_water * organic_nitrogen_benth : mole_concentration_of_organic_detritus_expressed_as_nitrogen_in_benthos Les paramètres diagnostiques calculés sont les suivants : * XE : sea_surface_height_above_geoid * maximum_de_diat : maximum_diatom_mass_concentration_in_sea_water * maximum_de_dino : maximum_dinoflagellate_mass_concentration_in_sea_water * maximum_de_nano : maximum_nanoflagellate_mass_concentration_in_sea_water * grad_vert_salinite : maximum_vertical_gradient_of_sea_water_salinity * grad_vert_temp : maximum_vertical_gradient_of_sea_water_temperature * extinction_lumineuse : light_extinction_in_sea_water * prod_diat : cumulated_production_of_diatoms_in_sea_water_column_expressed_in_carbon * prod_dino : cumulated_production_of_dinoflagellates_in_sea_water_column_expressed_in_carbon * prod_nano : cumulated_production_of_nanoflagellates_in_sea_water_column_expressed_in_carbon * chlorophylle_a : chlorophyll_mass_concentration_in_sea_water * prod_cumul_chloro : cumulated_total_production_in_sea_water_column_expressed_in_carbon * maximum_de_phaeocystis : maximum_phaeocystis_mass_concentration_in_sea_water * prod_phaeocystis : cumulated_production_of_phaeocystis_in_sea_water_column_expressed_in_carbon * oxygen_saturation : oxygen_saturation * ammoniumGIRON_tracer_sign: mole_concentration_of_ammonium_in_sea_waterGIRON_tracer_sign * ammoniumGIRON_tracer_age: mole_concentration_of_ammonium_in_sea_waterGIRON_tracer_age * nitrateGIRON_tracer_sign: mole_concentration_of_nitrate_in_sea_waterGIRON_tracer_sign * nitrateGIRON_tracer_age: mole_concentration_of_nitrate_in_sea_waterGIRON_tracer_age * nanopicoplankton_nitrogenGIRON_tracer_sign: mole_concentration_of_nanoplankton_expressed_as_nitrogen_in_sea_waterGIRON_tracer_sign * nanopicoplankton_nitrogenGIRON_tracer_age: mole_concentration_of_nanoplankton_expressed_as_nitrogen_in_sea_waterGIRON_tracer_age * diatom_nitrogenGIRON_tracer_sign: mole_concentration_of_diatoms_expressed_as_nitrogen_in_sea_waterGIRON_tracer_sign * diatom_nitrogenGIRON_tracer_age: mole_concentration_of_diatoms_expressed_as_nitrogen_in_sea_waterGIRON_tracer_age * dinoflagellate_nitrogenGIRON_tracer_sign: mole_concentration_of_dinoflagellates_expressed_as_nitrogen_in_sea_waterGIRON_tracer_sign * dinoflagellate_nitrogenGIRON_tracer_age: mole_concentration_of_dinoflagellates_expressed_as_nitrogen_in_sea_waterGIRON_tracer_age * microzooplankton_nitrogenGIRON_tracer_sign: mole_concentration_of_microzooplankton_expressed_as_nitrogen_in_sea_waterGIRON_tracer_sign * microzooplankton_nitrogenGIRON_tracer_age: mole_concentration_of_microzooplankton_expressed_as_nitrogen_in_sea_waterGIRON_tracer_age * mesozooplankton_nitrogenGIRON_tracer_sign: mole_concentration_of_mesozooplankton_expressed_as_nitrogen_in_sea_waterGIRON_tracer_sign * mesozooplankton_nitrogenGIRON_tracer_age: mole_concentration_of_mesozooplankton_expressed_as_nitrogen_in_sea_waterGIRON_tracer_age * detrital_nitrogenGIRON_tracer_sign: mole_concentration_of_organic_detritus_expressed_as_nitrogen_in_sea_waterGIRON_tracer_sign * detrital_nitrogenGIRON_tracer_age: mole_concentration_of_organic_detritus_expressed_as_nitrogen_in_sea_waterGIRON_tracer_age * colonial_phaeocystis_nitrogenGIRON_tracer_sign: mole_concentration_of_colonial_phaeocystis_expressed_as_nitrogen_in_sea_waterGIRON_tracer_sign * colonial_phaeocystis_nitrogenGIRON_tracer_age: mole_concentration_of_colonial_phaeocystis_expressed_as_nitrogen_in_sea_waterGIRON_tracer_age * phaeocystis_cells_nitrogenGIRON_tracer_sign: mole_concentration_of_phaeocystis_cells_expressed_as_nitrogen_in_sea_waterGIRON_tracer_sign * phaeocystis_cells_nitrogenGIRON_tracer_age: mole_concentration_of_phaeocystis_cells_expressed_as_nitrogen_in_sea_waterGIRON_tracer_age * organic_nitrogen_benthGIRON_tracer_sign: mole_concentration_of_organic_detritus_expressed_as_nitrogen_in_benthosGIRON_tracer_sign * organic_nitrogen_benthGIRON_tracer_age: mole_concentration_of_organic_detritus_expressed_as_nitrogen_in_benthosGIRON_tracer_age * phytoplankton_sign_N_GIRON: nitrogen_fraction_in_phytoplankton_from_source_GIRON * phytoplankton_age_N_GIRON: age_of_nitrogen_fraction_in_phytoplankton_from_source_GIRON * ammoniumLOIRE_tracer_sign: mole_concentration_of_ammonium_in_sea_waterLOIRE_tracer_sign * ammoniumLOIRE_tracer_age: mole_concentration_of_ammonium_in_sea_waterLOIRE_tracer_age * nitrateLOIRE_tracer_sign: mole_concentration_of_nitrate_in_sea_waterLOIRE_tracer_sign * nitrateLOIRE_tracer_age: mole_concentration_of_nitrate_in_sea_waterLOIRE_tracer_age * nanopicoplankton_nitrogenLOIRE_tracer_sign: mole_concentration_of_nanoplankton_expressed_as_nitrogen_in_sea_waterLOIRE_tracer_sign * nanopicoplankton_nitrogenLOIRE_tracer_age: mole_concentration_of_nanoplankton_expressed_as_nitrogen_in_sea_waterLOIRE_tracer_age * diatom_nitrogenLOIRE_tracer_sign: mole_concentration_of_diatoms_expressed_as_nitrogen_in_sea_waterLOIRE_tracer_sign * diatom_nitrogenLOIRE_tracer_age: mole_concentration_of_diatoms_expressed_as_nitrogen_in_sea_waterLOIRE_tracer_age * dinoflagellate_nitrogenLOIRE_tracer_sign: mole_concentration_of_dinoflagellates_expressed_as_nitrogen_in_sea_waterLOIRE_tracer_sign * dinoflagellate_nitrogenLOIRE_tracer_age: mole_concentration_of_dinoflagellates_expressed_as_nitrogen_in_sea_waterLOIRE_tracer_age * microzooplankton_nitrogenLOIRE_tracer_sign: mole_concentration_of_microzooplankton_expressed_as_nitrogen_in_sea_waterLOIRE_tracer_sign * microzooplankton_nitrogenLOIRE_tracer_age: mole_concentration_of_microzooplankton_expressed_as_nitrogen_in_sea_waterLOIRE_tracer_age * mesozooplankton_nitrogenLOIRE_tracer_sign: mole_concentration_of_mesozooplankton_expressed_as_nitrogen_in_sea_waterLOIRE_tracer_sign * mesozooplankton_nitrogenLOIRE_tracer_age: mole_concentration_of_mesozooplankton_expressed_as_nitrogen_in_sea_waterLOIRE_tracer_age * detrital_nitrogenLOIRE_tracer_sign: mole_concentration_of_organic_detritus_expressed_as_nitrogen_in_sea_waterLOIRE_tracer_sign * detrital_nitrogenLOIRE_tracer_age: mole_concentration_of_organic_detritus_expressed_as_nitrogen_in_sea_waterLOIRE_tracer_age * colonial_phaeocystis_nitrogenLOIRE_tracer_sign: mole_concentration_of_colonial_phaeocystis_expressed_as_nitrogen_in_sea_waterLOIRE_tracer_sign * colonial_phaeocystis_nitrogenLOIRE_tracer_age: mole_concentration_of_colonial_phaeocystis_expressed_as_nitrogen_in_sea_waterLOIRE_tracer_age * phaeocystis_cells_nitrogenLOIRE_tracer_sign: mole_concentration_of_phaeocystis_cells_expressed_as_nitrogen_in_sea_waterLOIRE_tracer_sign * phaeocystis_cells_nitrogenLOIRE_tracer_age: mole_concentration_of_phaeocystis_cells_expressed_as_nitrogen_in_sea_waterLOIRE_tracer_age * organic_nitrogen_benthLOIRE_tracer_sign: mole_concentration_of_organic_detritus_expressed_as_nitrogen_in_benthosLOIRE_tracer_sign * organic_nitrogen_benthLOIRE_tracer_age: mole_concentration_of_organic_detritus_expressed_as_nitrogen_in_benthosLOIRE_tracer_age * phytoplankton_sign_N_LOIRE: nitrogen_fraction_in_phytoplankton_from_source_LOIRE * phytoplankton_age_N_LOIRE: age_of_nitrogen_fraction_in_phytoplankton_from_source_LOIRE * ammoniumSEINE_tracer_sign: mole_concentration_of_ammonium_in_sea_waterSEINE_tracer_sign * ammoniumSEINE_tracer_age: mole_concentration_of_ammonium_in_sea_waterSEINE_tracer_age * nitrateSEINE_tracer_sign: mole_concentration_of_nitrate_in_sea_waterSEINE_tracer_sign * nitrateSEINE_tracer_age: mole_concentration_of_nitrate_in_sea_waterSEINE_tracer_age * nanopicoplankton_nitrogenSEINE_tracer_sign: mole_concentration_of_nanoplankton_expressed_as_nitrogen_in_sea_waterSEINE_tracer_sign * nanopicoplankton_nitrogenSEINE_tracer_age: mole_concentration_of_nanoplankton_expressed_as_nitrogen_in_sea_waterSEINE_tracer_age * diatom_nitrogenSEINE_tracer_sign: mole_concentration_of_diatoms_expressed_as_nitrogen_in_sea_waterSEINE_tracer_sign * diatom_nitrogenSEINE_tracer_age: mole_concentration_of_diatoms_expressed_as_nitrogen_in_sea_waterSEINE_tracer_age * dinoflagellate_nitrogenSEINE_tracer_sign: mole_concentration_of_dinoflagellates_expressed_as_nitrogen_in_sea_waterSEINE_tracer_sign * dinoflagellate_nitrogenSEINE_tracer_age: mole_concentration_of_dinoflagellates_expressed_as_nitrogen_in_sea_waterSEINE_tracer_age * microzooplankton_nitrogenSEINE_tracer_sign: mole_concentration_of_microzooplankton_expressed_as_nitrogen_in_sea_waterSEINE_tracer_sign * microzooplankton_nitrogenSEINE_tracer_age: mole_concentration_of_microzooplankton_expressed_as_nitrogen_in_sea_waterSEINE_tracer_age * mesozooplankton_nitrogenSEINE_tracer_sign: mole_concentration_of_mesozooplankton_expressed_as_nitrogen_in_sea_waterSEINE_tracer_sign * mesozooplankton_nitrogenSEINE_tracer_age: mole_concentration_of_mesozooplankton_expressed_as_nitrogen_in_sea_waterSEINE_tracer_age * detrital_nitrogenSEINE_tracer_sign: mole_concentration_of_organic_detritus_expressed_as_nitrogen_in_sea_waterSEINE_tracer_sign * detrital_nitrogenSEINE_tracer_age: mole_concentration_of_organic_detritus_expressed_as_nitrogen_in_sea_waterSEINE_tracer_age * colonial_phaeocystis_nitrogenSEINE_tracer_sign: mole_concentration_of_colonial_phaeocystis_expressed_as_nitrogen_in_sea_waterSEINE_tracer_sign * colonial_phaeocystis_nitrogenSEINE_tracer_age: mole_concentration_of_colonial_phaeocystis_expressed_as_nitrogen_in_sea_waterSEINE_tracer_age * phaeocystis_cells_nitrogenSEINE_tracer_sign: mole_concentration_of_phaeocystis_cells_expressed_as_nitrogen_in_sea_waterSEINE_tracer_sign * phaeocystis_cells_nitrogenSEINE_tracer_age: mole_concentration_of_phaeocystis_cells_expressed_as_nitrogen_in_sea_waterSEINE_tracer_age * organic_nitrogen_benthSEINE_tracer_sign: mole_concentration_of_organic_detritus_expressed_as_nitrogen_in_benthosSEINE_tracer_sign * organic_nitrogen_benthSEINE_tracer_age: mole_concentration_of_organic_detritus_expressed_as_nitrogen_in_benthosSEINE_tracer_age * phytoplankton_sign_N_SEINE: nitrogen_fraction_in_phytoplankton_from_source_SEINE * phytoplankton_age_N_SEINE: age_of_nitrogen_fraction_in_phytoplankton_from_source_SEINE
-
-

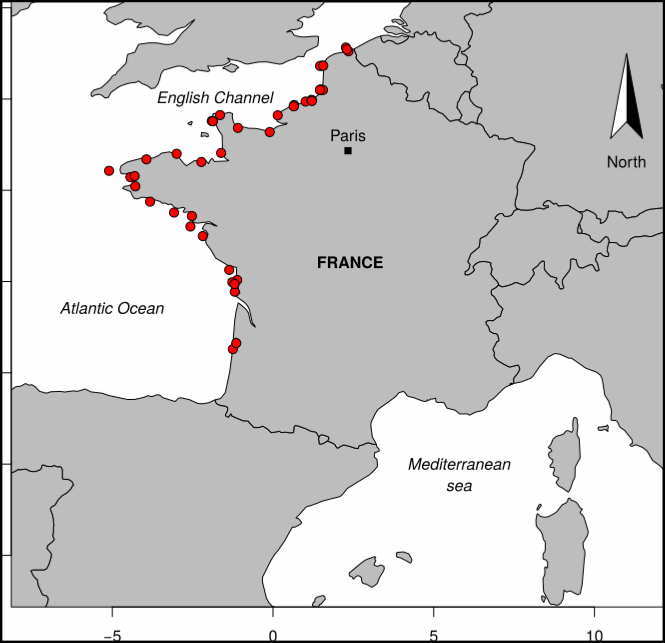
Plankton was sampled with a Continuous Underway Fish Egg Sampler (CUFES, 315µm mesh size) at 4 m below the surface, and a WP2 net (200µm mesh size) from 100m to the surface, or 5 m above the sea floor to the surface when the depth was < 100 m, in the Bay of Biscay. The full images were processed with the ZooCAM software and the embedded Matrox Imaging Library (Colas et a., 2018) which generated regions of interest (ROIs) around each individual object and a set of features measured on the object. The same objects were re-processed to compute features with the scikit-image library http://scikit-image.org. The 1, 286, 590 resulting objects were sorted by a limited number of operators, following a common taxonomic guide, into 93 taxa, using the web application EcoTaxa http://ecotaxa.obs-vlfr.fr. For the purpose of training machine learning classifiers, the images in each class were split into training, validation, and test sets, with proportions 70%, 15% and 15%. The archive contains : taxa.csv.gz Table of the classification of each object in the dataset, with columns : - objid : unique object identifier in EcoTaxa (integer number). - taxon_level1 : taxonomic name corresponding to the level 1 classification - lineage_level1 : taxonomic lineage corresponding to the level 1 classification - taxon_level2 : name of the taxon corresponding to the level 2 classification - plankton : if the object is a plankton or not (boolean) - set : class of the image corresponding to the taxon (train : training, val : validation, or test) - img_path : local path of the image corresponding to the taxon (of level 1), named according to the object id features_native.csv.gz Table of morphological features computed by ZooCAM. All features are computed on the object only, not the background. All area/length measures are in pixels. All grey levels are in encoded in 8 bits (0=black, 255=white). With columns : - area : object's surface - area_exc : object surface excluding white pixels - area_based_diameter : object's Area Based Diameter: 2 * (object_area/pi)^(1/2) - meangreyobjet : mean image grey level - modegreyobjet : modal object grey level - sigmagrey : object grey level standard deviation - mingrey : minimum object grey level - maxgrey : maximum object grey level - sumgrey : object grey level integrated density: object_mean*object_area - breadth : breadth of the object along the best fitting ellipsoid minor axis - length : breadth of the object along the best fitting ellipsoid majorr axis - elongation : elongation index: object_length/object_breadth - perim : object's perimeter - minferetdiam : minimum object's feret diameter - maxferetdiam : maximum object's feret diameter - meanferetdiam : average object's feret diameter - feretelongation : elongation index: object_maxferetdiam/object_minferetdiam - compactness : Isoperimetric quotient: the ration of the object's area to the area of a circle having the same perimeter - intercept0, intercept45 , intercept90, intercept135 : the number of times that a transition from background to foreground occurs a the angle 0ø, 45ø, 90ø and 135ø for the entire object - convexhullarea : area of the convex hull of the object - convexhullfillratio : ratio object_area/convexhullarea - convexperimeter : perimeter of the convex hull of the object - n_number_of_runs : number of horizontal strings of consecutive foreground pixels in the object - n_chained_pixels : number of chained pixels in the object - n_convex_hull_points : number of summits of the object's convex hull polygon - n_number_of_holes : number of holes (as closed white pixel area) in the object - roughness : measure of small scale variations of amplitude in the object's grey levels - rectangularity : ratio of the object's area over its best bounding rectangle's area - skewness : skewness of the object's grey level distribution - kurtosis : kurtosis of the object's grey level distribution - fractal_box : fractal dimension of the object's perimeter - hist25, hist50, hist75 : grey level value at quantile 0.25, 0.5 and 0.75 of the object's grey levels normalized cumulative histogram - valhist25, valhist50, valhist75 : sum of grey levels at quantile 0.25, 0.5 and 0.75 of the object's grey levels normalized cumulative histogram - nobj25, nobj50, nobj75 : number of objects after thresholding at the object_valhist25, object_valhist50 and object_valhist75 grey level - symetrieh :index of horizontal symmetry - symetriev : index of vertical symmetry - skelarea : area of the object skeleton - thick_r : maximum object's thickness/mean object's thickness - cdist : distance between the mass and the grey level object's centroids features_skimage.csv.gz Table of morphological features recomputed with skimage.measure.regionprops on the ROIs produced by ZooCAM. See http://scikit-image.org/docs/dev/api/skimage.measure.html#skimage.measure.regionprops for documentation. inventory.tsv Tree view of the taxonomy and number of images in each taxon, displayed as text. With columns : - lineage_level1 : taxonomic lineage corresponding to the level 1 classification - taxon_level1 : name of the taxon corresponding to the level 1 classification - n : number of objects in each taxon group map.png Map of the sampling locations, to give an idea of the diversity sampled in this dataset. imgs Directory containing images of each object, named according to the object id objid and sorted in subdirectories according to their taxon.
-
-

-

Coastal Surveillance Through Observation of Ocean Color (COASTlOOC) oceanographic expeditions were conducted in 1997 and 1998 to examine the relationship between the optical properties of seawater and related biological and chemical properties across the coastal-to-open ocean gradient in various European seas. A total of 379 stations were visited along the coasts of the Gulf of Lion in the Mediterranean Sea (n = 61), Adriatic Sea (n = 39), Baltic Sea (n = 57), North Sea (n = 99), English Channel (n = 85) and Atlantic Ocean (n = 38). Particular emphasis was dedicated to the collection of a comprehensive set of apparent (AOPs) and inherent (IOPs) optical properties to support the development of ocean color remote sensing algorithms. The data were collected in situ using traditional ship-based sampling, but also from a helicopter, which is a very efficient means for that type of coastal sampling. The dataset collected during the COASTlOOC campaigns is unique in that it is fully consistent in terms of operators, protocols, and instrumentation. This rich and historical dataset is still today frequently requested and used by other researchers. Therefore, we present the result of an effort to compile and standardize a dataset which will facilitate its use in future development and evaluation of new bio-optical models adapted for optically-complex waters.
-
 Catalogue PIGMA
Catalogue PIGMA