Bioinformatics
Type of resources
Available actions
Topics
Keywords
Contact for the resource
Provided by
Years
Formats
Representation types
-

This dataset consists of metatranscriptomic sequencing reads corresponding to coastal micro-eukaryote communities sampled in Western Europe in 2018 and 2019.
-
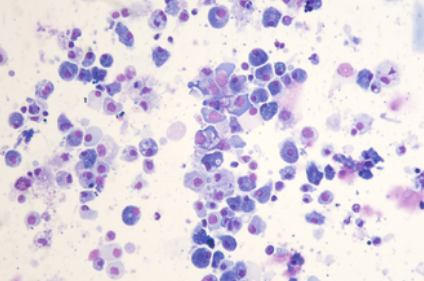
scRNA-seq reads from a Pacific oyster (Crassostrea gigas) hemocyte preparation. Hemocytes were isolated from a unique immunologically naive animal (Ifremer Standardized Animal, 18 months) and single-cell drop-seq technology was applied to 3,000 individual hemocytes.
-

DNA sequencing of Crassostrea gigas Pacific oyster spat infected in the wild with OsHV-1 virus in 4 French oyster basins (Marennes Oleron Bay, Arcachon bay, Rade de Brest and Thau lagoon).
-

Vibrio bacteria sampled from juvenile oysters and seawater collected in Thau Lagoon (Languedoc-Roussillon, France) in October 2015 during a mortality event were genotyped using hsp60, rctB, topA and mreB protein-coding genes
-

Raw reads for the assembly of Gambusia holbrooki genome.
-

Sequenced samples are city center wastewater sampled by passive samplers. Variants are identified by Illumina Miseq sequencing.
-

Land-sea continuum microbiome analyses in 4 coastal French sites and in oysters aimed at evaluating human impact on coastal ecosystems and new potentiel microbiological sanitary risks.
-

WGS of SARS-CoV-2 by Oxford Nanopore Technology from raw wastewater samples collected in France, 2020-2021
-

160 whole genomes sequences obtained from 160 individual fish samples representing about 100 different species present in Gulf of Lion, and bay of Biscay.
-

Dart Seq data gathered on Blue Shark in the framework of the PSTBS-IO project supported by funding from FAO, CSIRO Oceans and Atmosphere, AZTI Tecnalia, Institut de recherche pour le développement (IRD), and Research Institute for Tuna Fisheries (RITF) and financial assistance of the European Union (GCP/INT/233/EC – Population structure of IOTC species in the Indian Ocean), and POPSIZE project supported by FEAMP (2014-2020 UE N°508/2014), and Institut français de recherche pour l'Exploitation de la mer (Ifremer).
 Catalogue PIGMA
Catalogue PIGMA