2022
Type of resources
Available actions
Topics
Keywords
Contact for the resource
Provided by
Years
Formats
Representation types
Update frequencies
status
Service types
Scale
Resolution
-
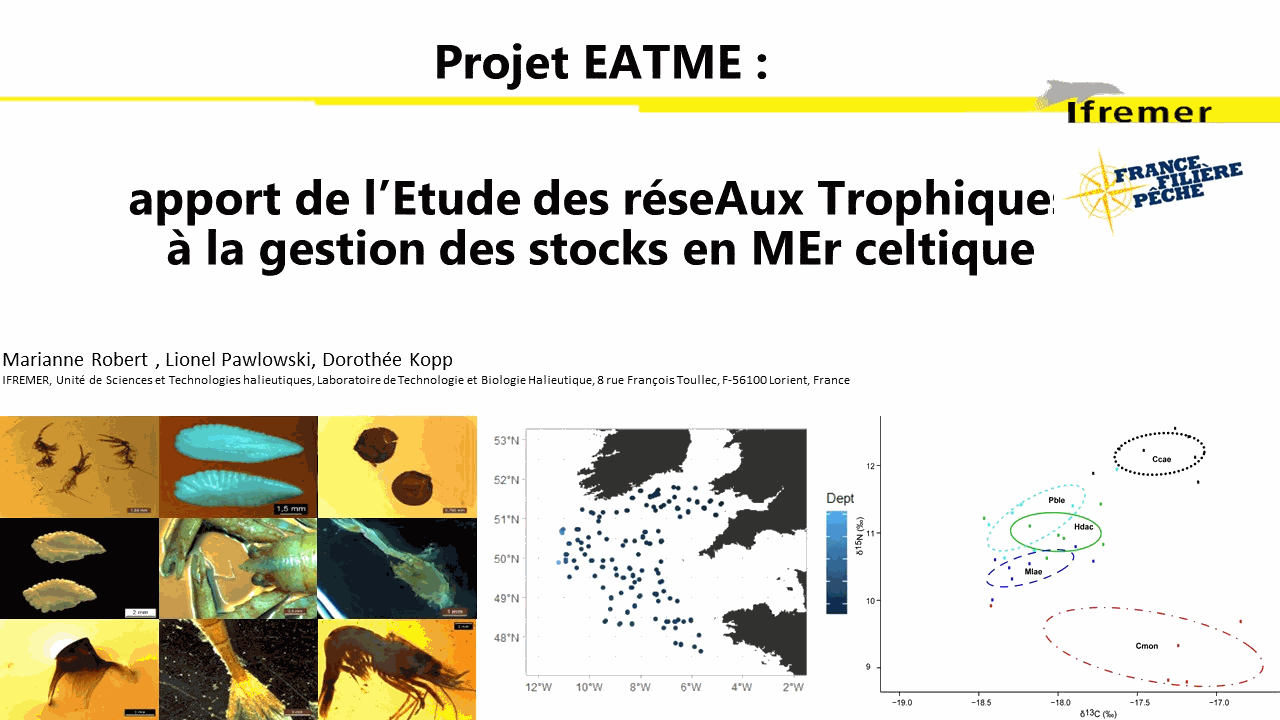
Understanding the dynamics of species interactions for food (prey-predator, competition for resources) and the functioning of trophic networks (dependence on trophic pathways, food chain flows, etc.) has become a thriving ecological research field in recent decades. This empirical knowledge is then used to develop population and ecosystem modelling approaches to support ecosystem-based management. The TrophicCS data set offers spatialized trophic information on a large spatial scale (the entire Celtic Sea continental shelf and upper slope) for a wide range of species. It combines ingested prey (gut content analysis) and a more integrated indicator of food sources (stable isotope analysis). A total of 1337 samples of large epifaunal invertebrates (bivalve mollusks and decapod crustaceans), zooplankton, fish and cephalopods, corresponding to 114 species, were collected and analyzed for stable isotope analysis of their carbon and nitrogen content. Sample size varied between taxa (from 1 to 52), with an average of 11.72 individuals sampled per species, and water depths ranged from 57 to 516 m. The gut contents of 1026 fish belonging to ten commercially important species: black anglerfish (Lophius budegassa), white anglerfish (Lophius piscatorius), blue whiting (Micromesistius poutassou), cod (Gadus morhua), haddock (Melanogrammus aeglefinus), hake (Merluccius merluccius), megrim (Lepidorhombus whiffiagonis), plaice (Pleuronectes platessa), sole (Solea solea) and whiting (Merlangius merlangus) were analyzed. The stomach content data set contains the occurrence of prey in stomach, identified to the lowest taxonomic level possible. To consider potential ontogenetic diet changes, a large size range was sampled. The TrophicCS data set was used to improve understanding of trophic relationships and ecosystem functioning in the Celtic Sea. When you use the data in your publication, we request that you cite this data paper. If you use the present data set (TrophicCS) for the majority of the data analyzed in your study, you may wish to consider inviting at least one author of the core team of this data paper to become a collaborator /coauthor of your paper.
-
Global wave hindcast (1961-2020) at 1° resolution using CMIP6 wind and sea-ice forcings for ALL (historical), GHG (historical greenhouse-gas-only), AER (historical Anthropogenic-aerosol-only), NAT (historical natural only) scenario.
-

Raw reads for the assembly of Gambusia holbrooki genome.
-
The ESA Sea State Climate Change Initiative (CCI) project has produced global multi-sensor time-series of along-track satellite altimeter significant wave height data (referred to as Level 2P (L2P) data) with a particular focus for use in climate studies. This dataset contains the Version 3 Remote Sensing Significant Wave Height product, which provides along-track data at approximately 6 km spatial resolution, separated per satellite and pass, including all measurements with flags, corrections and extra parameters from other sources. These are expert products with rich content and no data loss. The altimeter data used in the Sea State CCI dataset v3 come from multiple satellite missions spanning from 2002 to 2022021 (Envisat, CryoSat-2, Jason-1, Jason-2, Jason-3, SARAL, Sentinel-3A), therefore spanning over a shorter time range than version 1.1. Unlike version 1.1, this version 3 involved a complete and consistent retracking of all the included altimeters. Many altimeters are bi-frequency (Ku-C or Ku-S) and only measurements in Ku band were used, for consistency reasons, being available on each altimeter but SARAL (Ka band).
-
This dataset contains the pictures used for morphometric measurements, as well as the elemental compositon and production rates data, of planktonic Rhizaria. Specimens were collected in the bay of Villefranche-sur-Mer in May 2019 and during the P2107 cruise in the California Current in July-August 2021. Analyses of the data can be found at https://github.com/MnnLgt/Elemental_composition_Rhizaria.
-
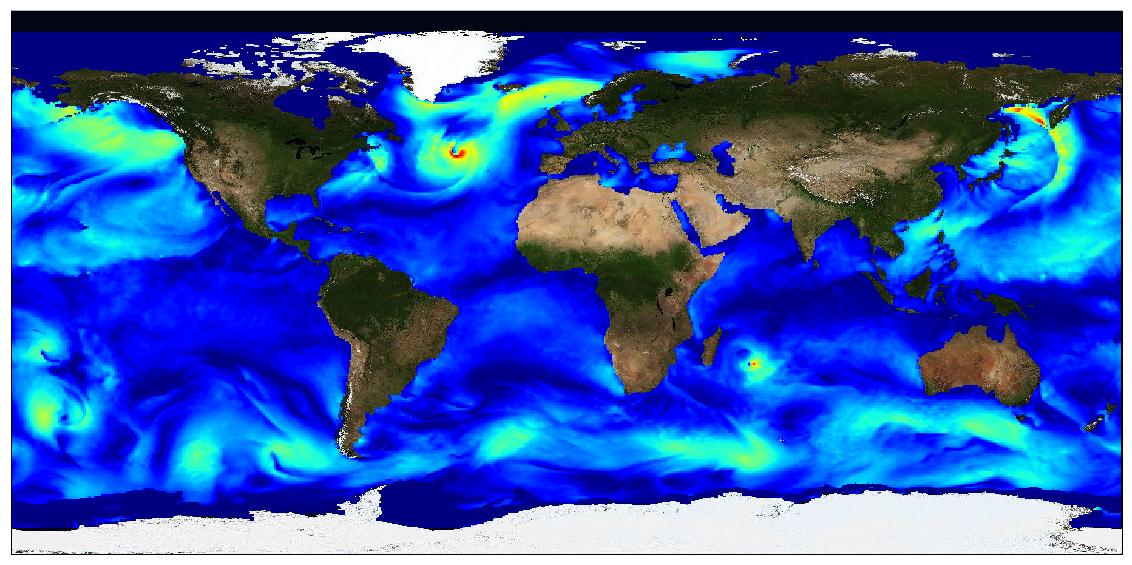
This dataset provides surface Stokes drift as retrieved from the wave energy spectrum computed by the spectral wave model WAVEWATCH-III (r), under NOAA license, discretized in wave numbers and directions and the water depth at each location. It is estimated at the sea surface and expressed in m.s-1. WAVEWATCH-III (r) model solves the random phase spectral action density balance equation for wavenumber-direction spectra. Please refer to the WAVEWATCH-III User Manual for fully detailed description of the wave model equations and numerical approaches. The data are available through HTTP and FTP; access to the data is free and open. In order to be informed about changes and to help us keep track of data usage, we encourage users to register at: https://forms.ifremer.fr/lops-siam/access-to-esa-world-ocean-circulation-project-data/ This dataset was generated by Ifremer / LOPS and is distributed by Ifremer / CERSAT in the frame of the World Ocean Circulation (WOC) project funded by the European Space Agency (ESA).
-
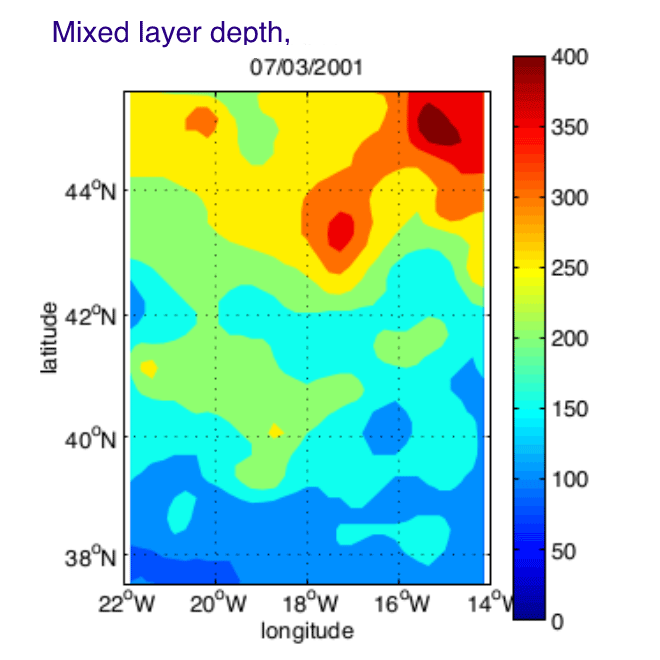
The Programme Ocean Multidisciplinaire Meso Echelle (POMME) was designed to describe and quantify the role of mesoscale processes in the subduction of mode waters in the Northeast Atlantic. Intensive situ measurements were maintained during 1 year (September 2000 - October 2001), over a 8 degrees square area centered on 18 degrees W, 42 degrees N. In order to synthesized the in-situ physical observations, and merge them with satellite altimetry and surface fluxes datasets, a simplified Kalman filter has been designed. Daily fields of temperature, salinity, and stream function were produced on a regular grid over a full seasonal cycle. We propose here the gridded fields (KA_ files) and the in-situ datasets used by the analysis (Data_ files).
-
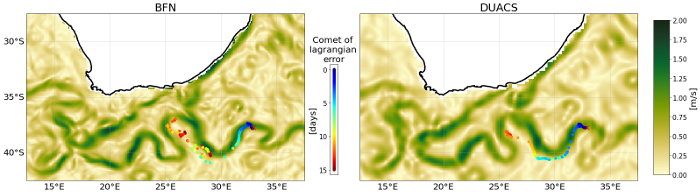
This dataset provides Level 4 total current including geostrophy and a data-driven approach for Ekman and near-inertial current, based on a convolution between drifter observation and wind history, to fit empirically a complex and time-lag dependant transfert function between ERA5 wind stress and current The data are available through HTTP and FTP; access to the data is free and open. In order to be informed about changes and to help us keep track of data usage, we encourage users to register at: https://forms.ifremer.fr/lops-siam/access-to-esa-world-ocean-circulation-project-data/ This dataset was generated by Datlas and is distributed by Ifremer / CERSAT in the frame of the World Ocean Circulation (WOC) project funded by the European Space Agency (ESA).
-
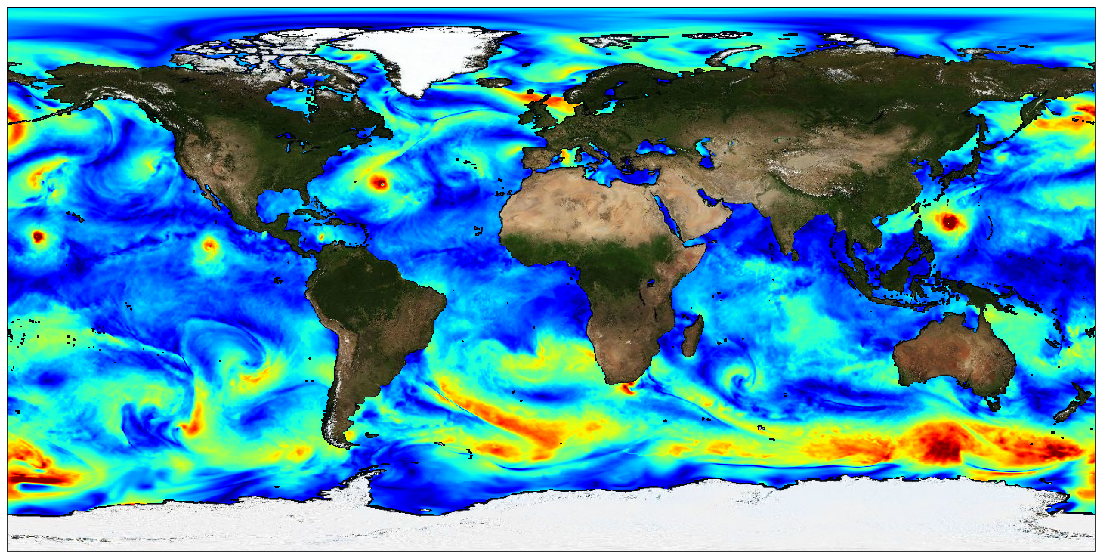
The Level 4 merged microwave wind product is a complete set of hourly global 10-m wind maps on a 0.25x0.25 degree latitude-longitude grid, spanning 1 Jan 2010 through the end of 2020. The product combines background neutral equivalent wind fields from ERA5, daily surface current fields from CMEMS, and stress equivalent winds obtained from several microwave passive and active sensors to produce hourly surface current relative stress equivalent wind analyses. The satellite winds include those from recently launched L-band passive sensors capable of measuring extreme winds in tropical cyclones, with little or no degradation from precipitation. All satellite winds used in the analyses have been recalibrated using a large set of collocated satellite-SFMR wind data in storm-centric coordinates. To maximize the use of the satellite microwave data, winds within a 24-hour window centered on the analysis time have been incorporated into each analysis. To accomodate the large time window, satellite wind speeds are transformed into deviations from ERA5 background wind speeds interpolated to the measurement times, and then an optical flow-based morphing technique is applied to these wind speed increments to propagate them from measurement to analysis time. These morphed wind speed increments are then added to the background wind speed at the analysis time to yield a set of total wind speeds fields for each sensor at the analysis time. These individual sensor wind speed fields are then combined with the background 10-m wind direction to yield vorticity and divergence fields for the individual sensor winds. From these, merged vorticity and divergence fields are computed as a weighted average of the individual vorticity and divergence fields. The final vector wind field is then obtained directly from these merged vorticity and divergence fields. Note that one consequence of producing the analyses in terms of vorticity and divergence is that there are no discontinuities in the wind speed fields at the (morphed) swath edges. There are two important points to be noted: the background ERA5 wind speed fields have been rescaled to be globally consistent with the recalibrated AMSR2 wind speeds. This rescaling involves a large increase in the ERA5 background winds beyond about 17 m/s. For example, an ERA5 10 m wind speed of 30 m/s is transformed into a wind speed of 41 m/s, and a wind speed of 34 m/s is transformed into a wind speed of about 48 m/s. Besides the current version of the product is calibrated for use within tropical cyclones and is not appropriate for use elsewhere. This dataset was produced in the frame of ESA MAXSS project. The primary objective of the ESA Marine Atmosphere eXtreme Satellite Synergy (MAXSS) project is to provide guidance and innovative methodologies to maximize the synergetic use of available Earth Observation data (satellite, in situ) to improve understanding about the multi-scale dynamical characteristics of extreme air-sea interaction.
-

We genotyped 1680 thornback ray Raja clavata sampled in the Bay of Biscay using a DNA chip described in Le Cam et al. (2019). After quality control 4604 SNPs were retained for identifying potential sex-linked SNPs using three methods: i) identification of excess of heterozygotes in one sex, ii) FST outlier analysis between the two sexes and iii) neuronal net modelling. Genotype coding: 0 homozygous for major allele, 1 heterozygous, 2 homozygous for minor allele. Flanking DNA sequences of SNPs identified with methods i) and ii) are also provided.
 Catalogue PIGMA
Catalogue PIGMA